Groups:
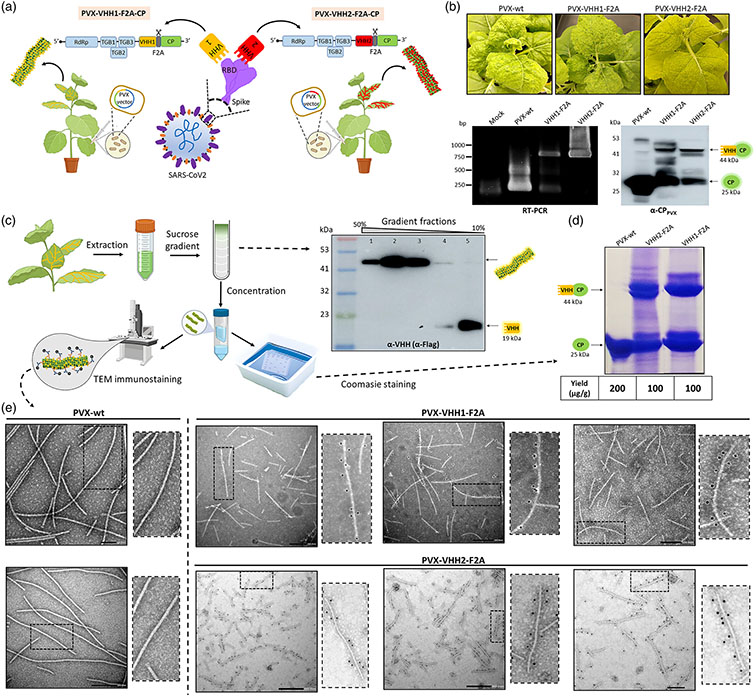
Plant Virus Biotechnology
José-Antonio Daròs IBMCP (CSIC-Universitat Politècnica de València)
Plants host a large variety of infectious agents that frequently cause damage to crops and natural ecosystems. The first goal of the research group is to understand the molecular mechanisms that underlie the interaction between plants and some of these pathogens, such as viruses and viroids. From this knowledge we expect to develop new biotechnological strategies for crop protection. In addition, we believe that we can take advantage of the remarkable biological properties of plant viruses and viroids by converting them into useful biotechnological tools. Our second goal is to develop systems to obtain products of interest, such as recombinant proteins, metabolites, RNAs or nanoparticles, in biofactory plants using conveniently engineered viruses and viroids. We envision a future in which cultivated plants will be the most reliable and sustainable source of food, feed, fibers, fuel, and pharmaceutical and chemical products for human kind.
Molecular Biology of Viral and Subviral Plant Pathogens
Carmen Hernández IBMCP (CSIC-Universitat Politècnica de València)
The group's work is fundamentally directed at the study of the factors that determine the infection/accumulation of a virus in a given host. As this accumulation will essentially be the result of the balance between viral multiplication and degradation, our attention is focused on the events that influence these two processes. Specifically, we are prioritizing the analysis of the mechanisms of replication, transcription and translation of viral genomes as well as the strategies developed by the pathogen to suppress the host's defensive response based on RNA silencing. As models, we are using small viruses with an RNA genome, belonging to the Tombusviridae family, which are characterized by having a relatively simple genome, which facilitates experimental approaches. Briefly, the current lines of research would be: (1) Identification and characterization of structural elements of the viral RNA and protein factors (viral/cellular) involved in the regulation of virus replication, transcription and/or translation processes. (2) Study of the viral function of suppression of RNA silencing. (3) Analysis of the possible interference of viral infection with host silencing pathways.
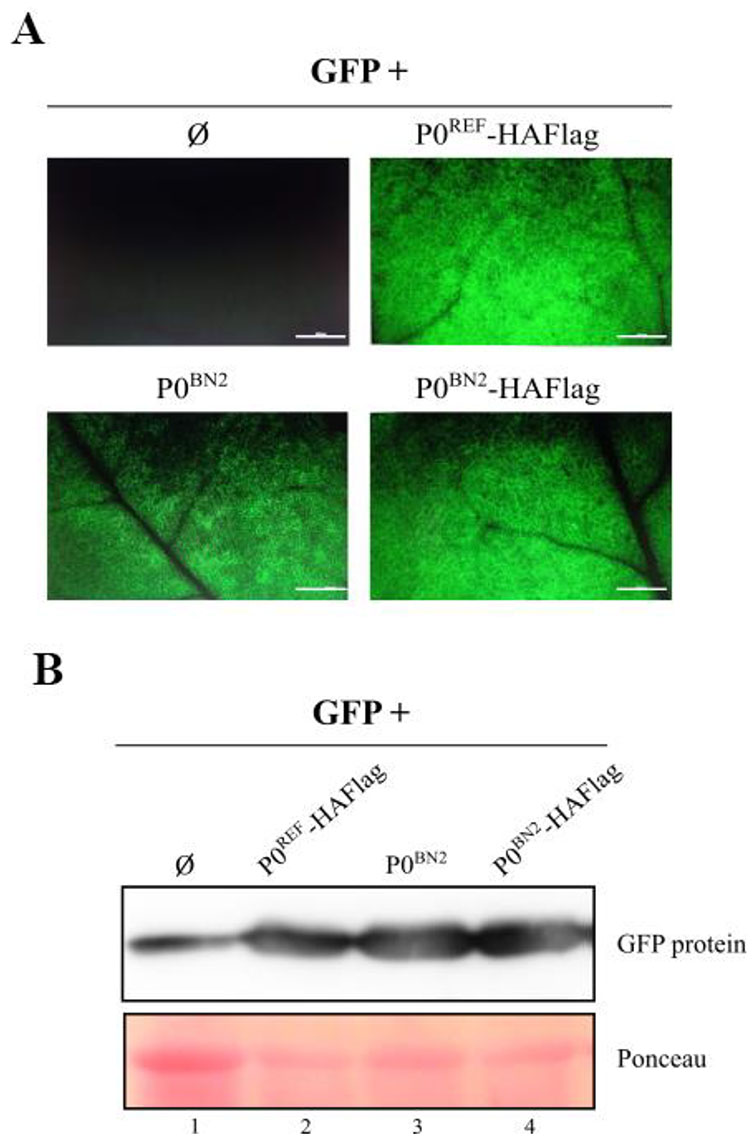
Evaluation of RNA silencing suppressor activity of the natural variant PeVYV-5 P0BN2.
BioSystems Design
Guillermo Rodrigo I2SysBio (CSIC-Universitat de València)
We engineer synthetic gene circuits aimed at (re-)programming living cells with repurposed and de novo designed regulatory elements. We are especially appealed by RNA-based control mechanisms. We develop computational models at different levels of intracellular organization (molecular, genetic, and genomic) to understand in a quantitative way the bases of gene regulation. We further engineer CRISPR-based detection methods and regulatory systems. In addition, we engineer genetic cassettes encoding viruses and virus-based nanoparticles following a sequence-structure-function paradigm. We also study how viruses evolve and examine virus-host interactions in terms of networks.
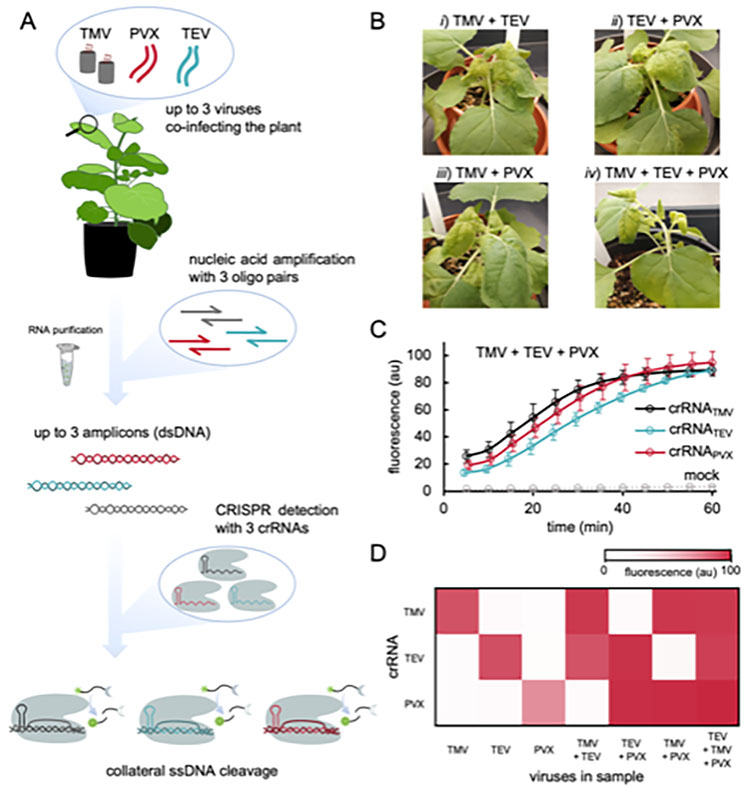
Multiplexed plant virus detection with a CRISPR-Cas12a-based assay.
Plants Genomics and Biotechnology
Diego Orzáez IBMCP (CSIC-Universitat Politècnica de València
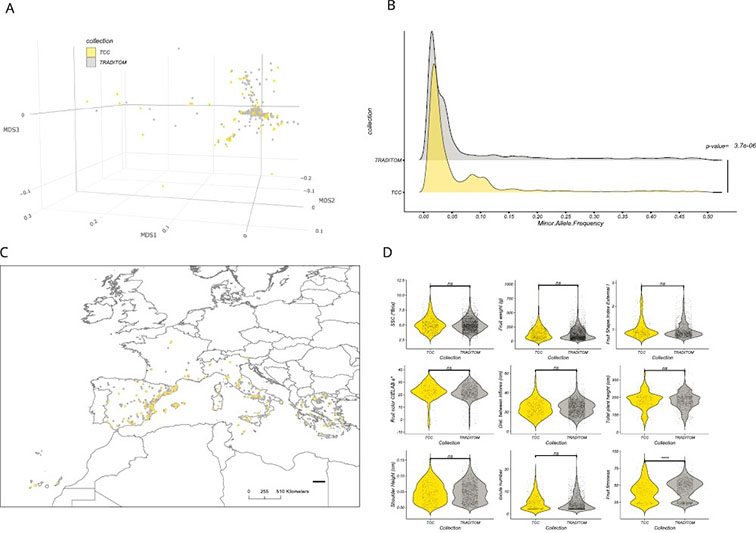
Our research group is interested in the design of innovative agricultural goods using genomics, biotechnology and synthetic biology tools. Our main experimental system is the tomato fruit, and our aim is to increase its added value with new shapes, colors, aromas and healthy properties. For this, we browse the natural variability of tomato-related wild species in order to identify loci conferring fruit quality traits. Once those loci are identified, we use marker-assisted breeding to transfer them to modern tomato cultivars. Moving beyond the limits imposed by sexual hybridization and natural variability, we also make use of biotechnology tools as multigene engineering and genetic transformation to bring new quality traits into the tomato genome. In a second line of research, we employ Synthetic Biology tools and principles for engineering plant biofactories of added-value products. In our lab, we have learnt how to build increasingly complex genetic instructions at the DNA level. By introducing these instructions into the genome of solanaceae species as tomato, tobacco and especially Nicotiana benthamiana (a tobacco relative), our goal is to re-direct the genetic program of the plant towards the synthesis of added-value products. Assisted by Agrobacterium tumefaciens and sometimes by viral vectors, we learn how to produce high levels of fine chemicals as antibodies, lectins, or snake anti-venoms in a short period of time. For more information, visit our group webpage at http://pgb.ibmcp.csic.es. Research interest: Molecular Breeding, Genetic Engineering and Synthetic Biology, Fruit Metabolomics and Molecular farming.
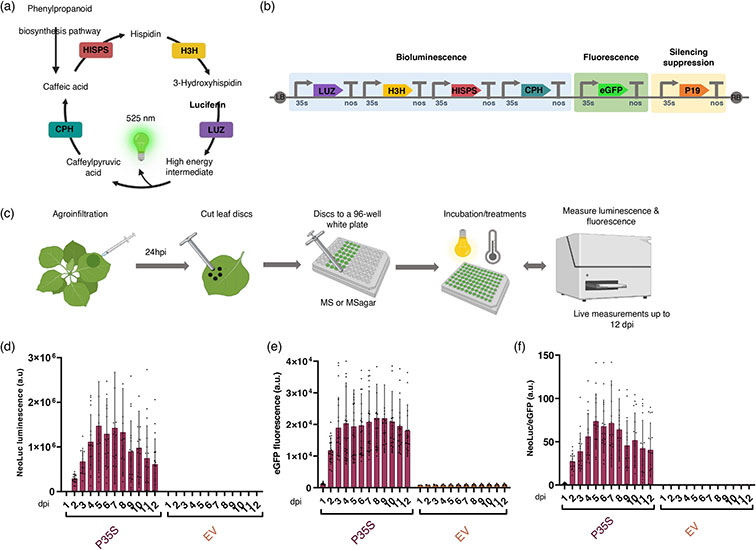
A quantitative bioluminescence reporter system for N. benthamiana.
Analysis and Applied Mathematics
Alberto Conejero IUMPA, Universitat Politècnica de València
I am interested in the mathematical modeling and data analysis of problems from any discipline, from physics or engineering, as well as from the social and life sciences. We find especially attractive those problems that allow them to be studied from the point of view of complex networks, graph theory, and applied mathematical analysis. (1) Analysis and Applied Mathematics. I am also interested in studying Linear Dynamics, Algebraic properties in linear spaces, and fractional calculus modelling. I am collaborating with M. Murillo-Arcila, A. Peris, and C. Lizama (GAFEVOL). (2) Biomedical data science. I am currently collaborating with the BDSLab (ITACA) on the following topics: Glioblastoma Modelling, Design of apps for collecting patient information to feed predictive models, and data quality of medical records repositories. (3) Diffusion processes and nonlinear phenomena in optics. I am interested in studying anomalous diffusion processes via machine learning. I am currently working on the regime change point detection of the process that involves. I am also interested in studying Fractional Calculus Modelling of such processes. I am collaborating with M.A. García March and C. Milián. (4) Mathematical Modeling in Synthetic Biology. I am working in plants with Diego Orzáez Lab (IBMCP) and at the cellular level with Guillermo Rodrigo (I2SysBio). (5) Mathematical Foundations of Quantum Computing. I am analyzing the structure and behavior of optimization algorithms that, in their configuration, use tensor products and their connections with Quantum Computing. I collaborate with Antonio Falcó (ESI Chair at Universidad Cardenal Herrera). (6) Data Science for Common and Social Good. I am a proud member of the Data Science against COVID-19 Task Force, led by Nuria Oliver (Ellis Alicante). We have specialized in outbreak modeling and predicting the evolution of COVID-19 pandemics via Recurrent Neural Networks. (7) Complex Networks Modelling. I am interested in applications to any discipline in network communications, image processing, linguistics, and many more. I am also working in data visualization of Science Maps in collaboration with E. Orduña and M. Rebollo. (8) Modelling in Industrial Engineering. I work in signal processing for the detection of failures in electrical machines & energy efficiency problems with Jose A. Antonino (Instituto Tecnología Energética).
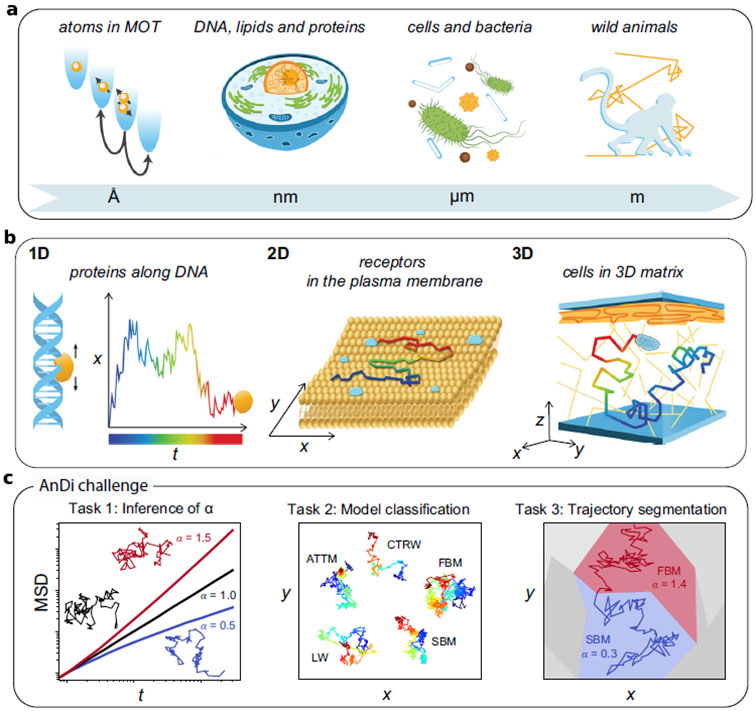
The AnDi challenge tasks and datasets
Functional Genomics and Biotechnology of ncRNAs
Marcos de la Peña IBMCP (CSIC-Universitat Politècnica de València
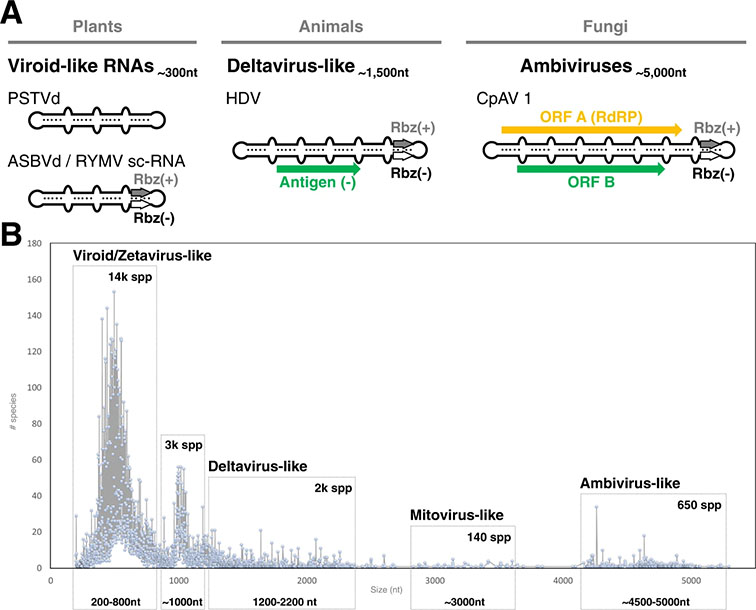
The development of massive nucleic acid sequencing techniques has allowed us to discover new ways and routes of regulation of gene expression of both living beings and viral agents. All this information is allowing us to better understand the high complexity of every biological entity on the planet, and makes us rethink both the classic concept of a gene and the function of each pair of bases in a genome. Within this revolution, the central role that RNA plays in a multitude of biological and infectious processes stands out. Our group is interested in finding and characterizing new biological functions for non-coding RNAs, focusing especially on their catalytic (ribozymes) and regulatory (riboregulators) capacities, as well as in biotechnologically exploiting any of the new RNA motifs detected. For all this, in the laboratory we use multidisciplinary approaches that combine Bioinformatics, Biochemistry and Molecular and Structural Biology, and fundamentally using plants as a reference experimental biological system. Among the specific lines of research that we currently follow, the following stand out: (1) Find and characterize the genome of all viruses and infectious RNAs on the planet through cloud computing approaches (de la Peña et al. Cells 2020; de la Peña et al. Virus Evol 2021; Edgar et al. Nature 2022). (2) Understand the functioning and biological role of circular RNAs with ribozymes encoded in plant and animal genomes (Cervera et al. Genome Biol 2016; de la Peña and Cevera. RNA Biol 2017; Cervera and de la Peña, Nucl Acids Res 2020). (3) Bioinformatics exploration and characterization of new catalytic and regulatory RNA motifs (de la Peña et al. Molecules 2017). (4) Functional characterization of small ribozymes associated with retrotransposons (Cervera and de la Peña, Mol Biol Evol 2014). (5) Deciphering the role of ribozymes in the human genome conserved in introns of antioncogenes and tumor antigens (de la Peña, García-Robles. EMBO Rep 2010; de la Peña et al. Biol Chem 2012; de la Peña, Ribozymes-Chapter 11 2021). (6) Development of possible biotechnological tools based on circular RNAs with ribozymes for gene therapies (de la Peña. Adv Exp Medicine Biology 2018).
Plants Genomics and Biotechnology
Antonio Granell IBMCP (CSIC-Universitat Politècnica de València
Our research group is interested in the design of innovative agricultural goods using genomics, biotechnology and synthetic biology tools. Our main experimental system is the tomato fruit, and our aim is to increase its added value with new shapes, colors, aromas and healthy properties. For this, we browse the natural variability of tomato-related wild species in order to identify loci conferring fruit quality traits. Once those loci are identified, we use marker-assisted breeding to transfer them to modern tomato cultivars. Moving beyond the limits imposed by sexual hybridization and natural variability, we also make use of biotechnology tools as multigene engineering and genetic transformation to bring new quality traits into the tomato genome. In a second line of research, we employ Synthetic Biology tools and principles for engineering plant biofactories of added-value products. In our lab, we have learnt how to build increasingly complex genetic instructions at the DNA level. By introducing these instructions into the genome of solanaceae species as tomato, tobacco and especially Nicotiana benthamiana (a tobacco relative), our goal is to re-direct the genetic program of the plant towards the synthesis of added-value products. Assisted by Agrobacterium tumefaciens and sometimes by viral vectors, we learn how to produce high levels of fine chemicals as antibodies, lectins, or snake anti-venoms in a short period of time. For more information, visit our group webpage at http://pgb.ibmcp.csic.es. Research interest: Molecular Breeding, Genetic Engineering and Synthetic Biology, Fruit Metabolomics and Molecular farming.